About
This app visualizes cell meta data, gene expression and motif activity information from scRNA and scATAC data in each inferred developmental time window. For additional data access and download please check out our resources page. For further detail please check out the manuscript.
These visualizations were built using an adapted version of the code used to visualize our other cell atlases. Please allow for additional load time for the initial loading and initial gene expression or motif activity queries. Contacts regarding this app can be sent to the Github page.
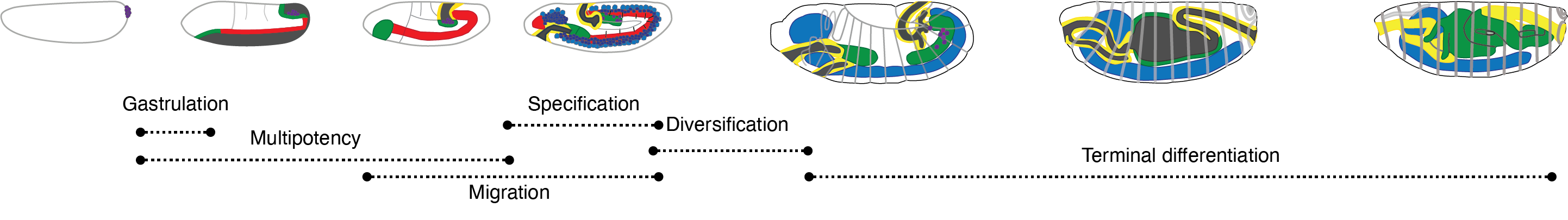
Drosophila melanogaster is a powerful, long-standing model for metazoan development and gene regulation. We profiled chromatin accessibility in almost one million, and gene expression in half a million, nuclei from eleven tightly staged, overlapping windows of its embryogenesis. Leveraging the asynchronicity of embryos within each collection, we developed a statistical model to more precisely estimate the age of each nucleus, resulting in continuous views of molecular and cellular transitions throughout the majority of embryonic development. From these data, we identify cell types, infer their developmental relationships, and link cell type-specific changes to dynamic changes in enhancer usage, transcription factor (TF) expression and the accessibility of TFs' cognate motifs. Looking forward, this strategy may facilitate future investigations of gene regulation in vivo at arbitrarily high temporal resolution.