About
In the mutant mouse cell atlas (MMCA) we profiled single cell transcriptomes in over 100 mouse embryos from 22 mutant strains and their corresponding wildtypes at the stage of embryonic day 13.5. The central aim of this project was to establish a platform for digital phenotyping of mouse mutants ranging in phenotype severity using sciRNA-seq3. In our manuscript titled "Single-cell, whole-embryo phenotyping of mammalian developmental disorders" (link) we have identified pleiotropic phenotypes affecting multiple trajectories within one mutant as well as subtle, single trajectory changes, demonstrating the capability of single cell technologies and our developed analytical tools to effectively detect and identify disease related changes within mouse development. On this website, we want to make our dataset available for further interactive exploration.
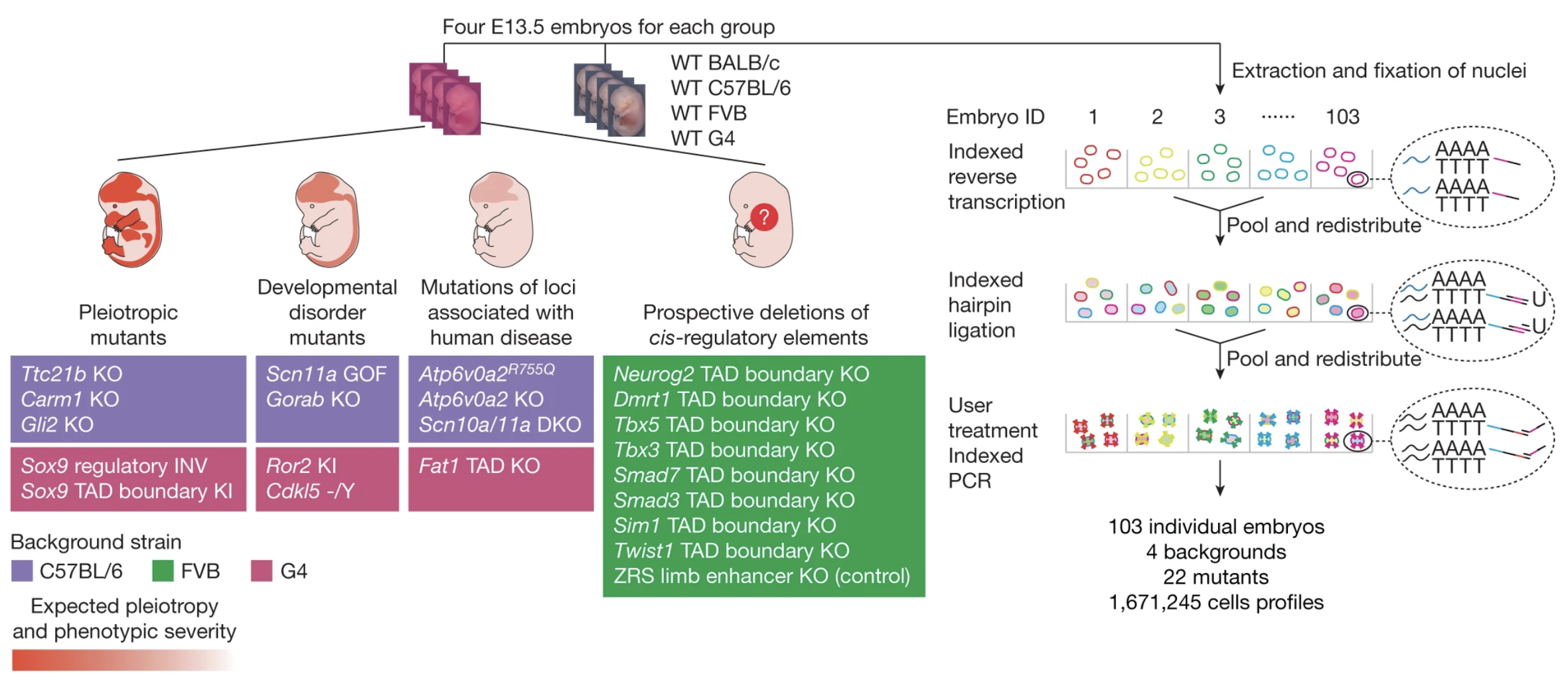
This app visualizes the single cell gene expression and metadata from mutant and wild-type mice. Each cell's embedding coordinates were determined using the UMAP algorithm. This resource was made in conjunction with a published scientific article. For further detail and in-depth analysis, please check out the paper.