Mouse sci-ATAC-seq Atlas
An atlas of chromatin accessibility in mouse at single cell resolution.
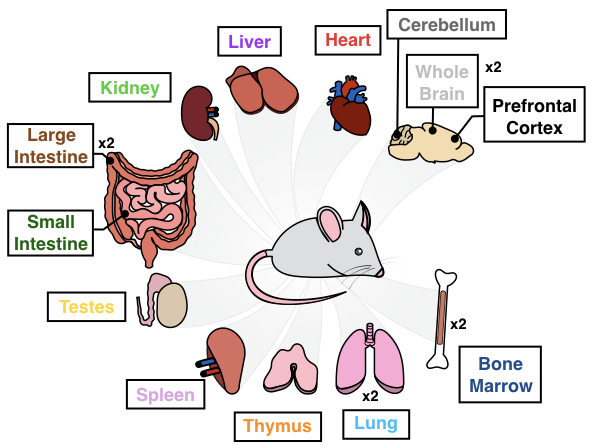
In our manuscript (Cusanovich and Hill, et al.), we report single cell measurements of chromatin accessibility for 17 samples spanning 13 different tissues in 8-week old mice (Mus. musculus).
Downloads
We have made our primary and secondary data available for the research community along with extensive documentation.
Getting Started Tutorial
We've put together tutorials on how to get started with analysis of sci-ATAC-seq data.
Visualizations
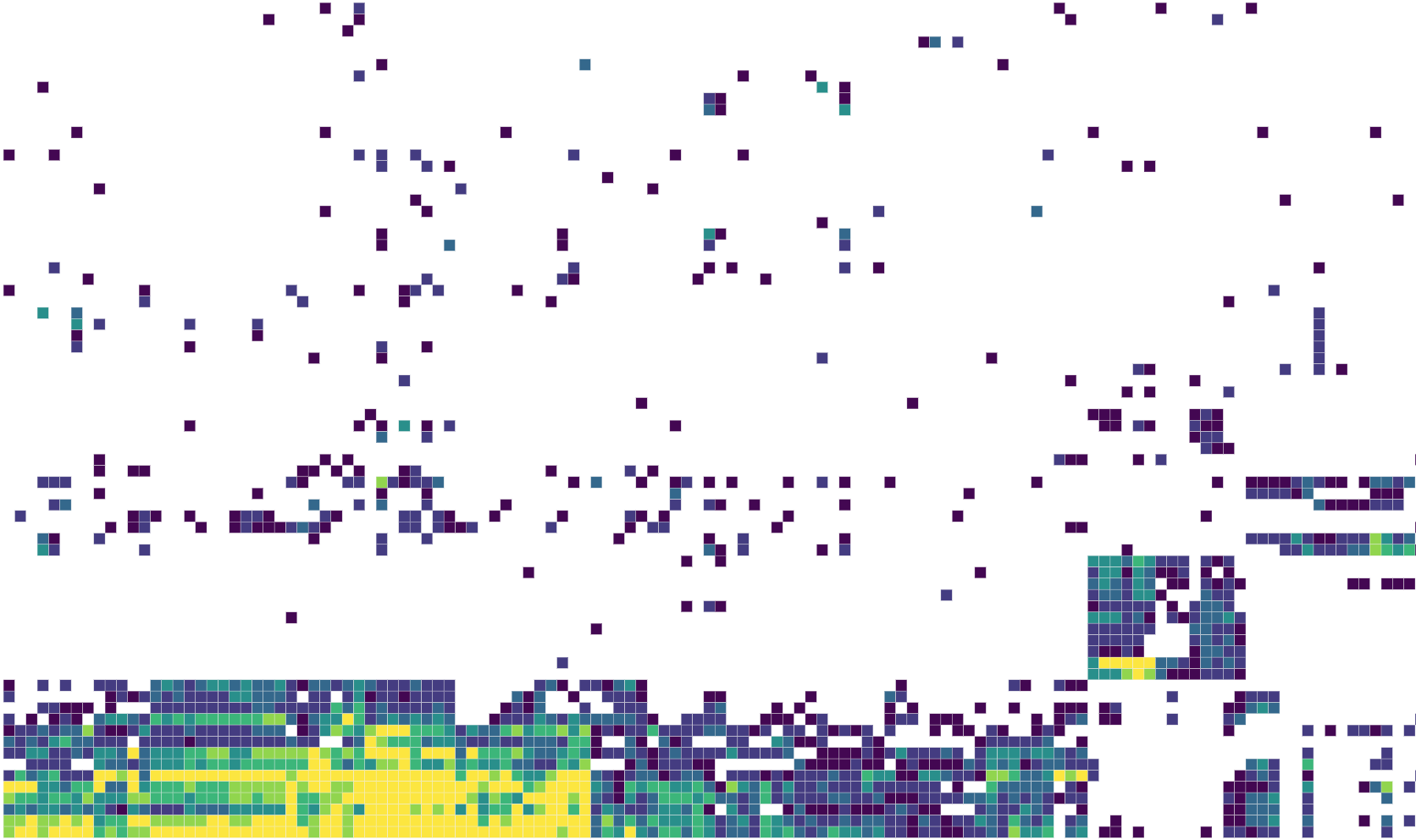
We have a few interactive visualization tools in progress to explore some of our results. Currently we have an interactive version of our figure on cell-type specific heritability enrichments for UKBB traits available as an example.
What is sci-ATAC-seq?
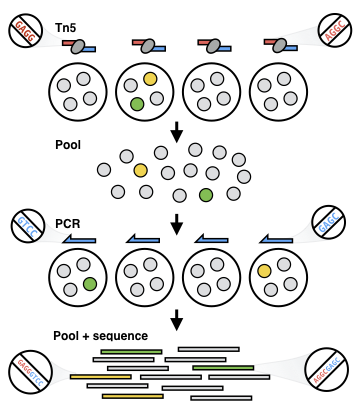
To generate these data we have used a technique we developed called sci-ATAC-seq (Cusanovich et al., Science 2015).
sci-ATAC-seq uses a paradigm called combinatorial indexing, where nucelic acids from cells are labeled with unique combinations of barcodes via multiple rounds of split-pool barcoding.